Collaborators: Fridtjof Brauns, M. Cristina Marchetti
Biological information is often discrete: cell positions, velocities, orientations, tractions. To create continuum fields out of this data many coarsegraining approaches are used, the most common among those are based on boxes. The field of view is divided into boxes whose size is inversely proportional to the density of data and quantities are calculated as averages within those boxes. This method suffers disadvantages when the data density is highly inhomogeneous: since a box size chosen according to average density will prove too large for dense areas and too small for sparser ones. We introduce a coarsegraining method that is independent of a coarsegraining grid and is based on the popular k-nearest-neighbor method used in data science. The schematic (taken from our preprint) is provided below. At any point, one can find the value of the coarsegrained field by averaging the data from k-nearest datapoints.
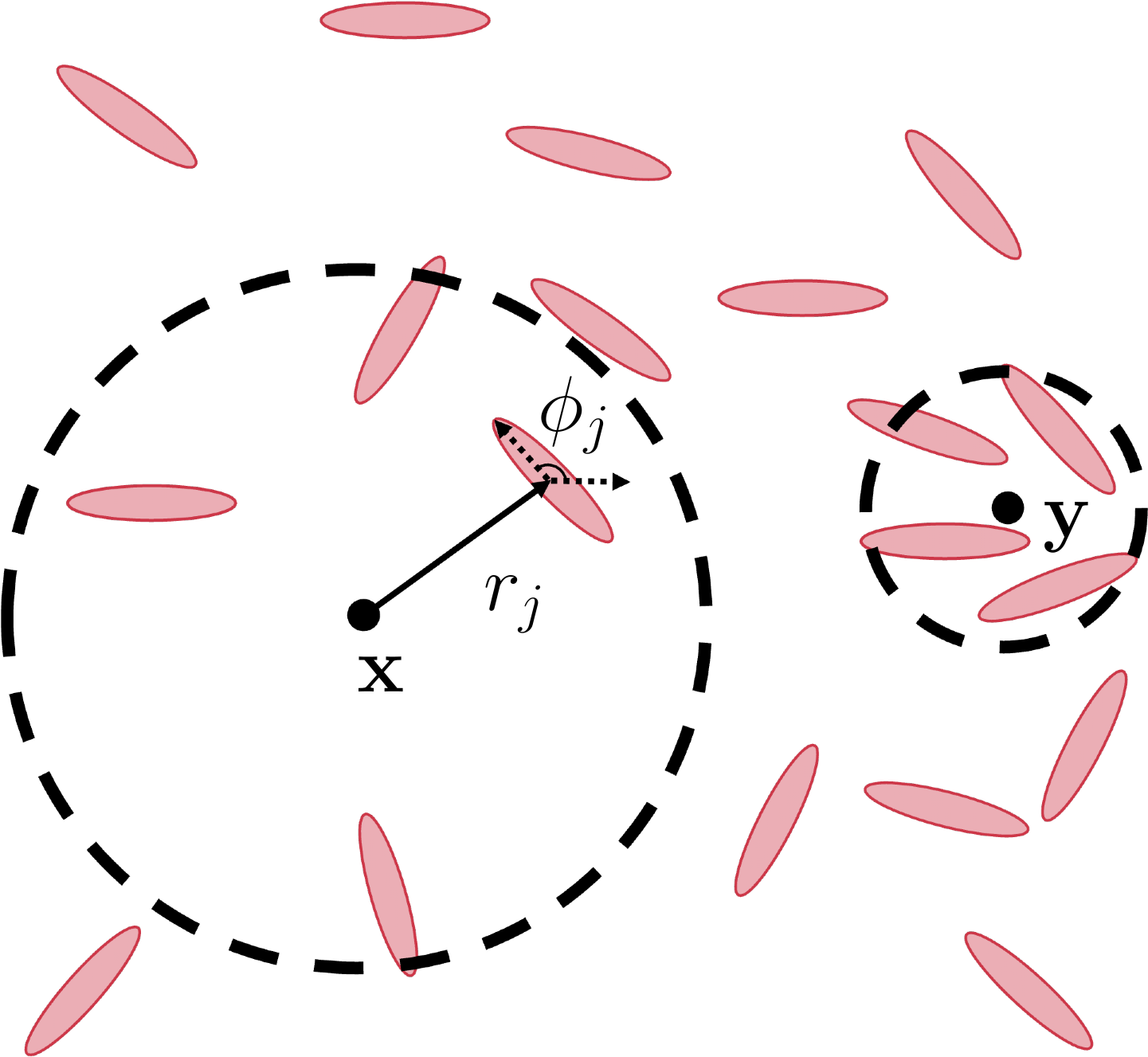
This method is akin to having a box-averaging method where the box size can vary depending on the local density of datapoints. We are in process of benchmarking the process on various discrete data. It is well known that the kNN method when used to approximate probability distribution functions underperforms when estimating distributions that are long-tailed. To counteract that, we update our method into a dynamic kNN method inspired by recent advances in data science.